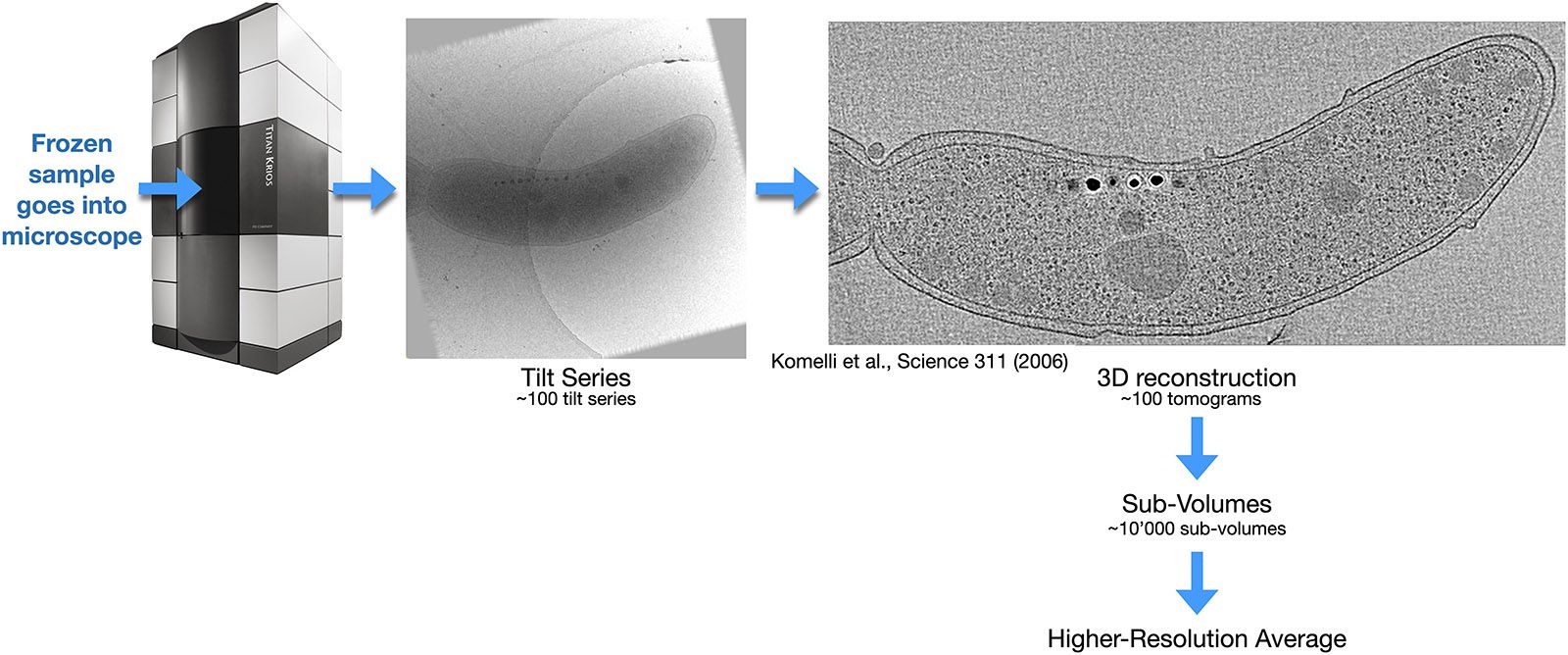
In electron tomography, each object of interest in the sample is imaged horizontally and at various angles to the electron beam. The recorded series of images from different tilt angles is then called a "tilt series". The increase in thickness to the electron beam of the tilted sample limits the possible angular range to ±60°. Computer image processing of the images gives a 3D reconstruction of the sample, yet a lower resolution, because individual movements of sub-sections of the sample during recording of the tilt series cannot be compensated at this step (it will be compensated later, see below). A first 3D reconstruction from a tilt series is therefore often limited to 2nm 3D resolution. Because of the limited tilt range, the whole 3D Fourier space cannot be sampled, so that the resolution in the z direction is lower than in the X and Y directions.
Recorded tilt series and their first 3D reconstruction can then be inspected manually or by computer software, to identify regions of interest. If for example the several ribosomes in a cell would be the objects of interest, then the X/Y/Y coordinates of each ribosome could be detected. If software then extracts from each two-dimensional image from the tilt series the image segment that belongs to a certain ribosome, then for each ribosome the computer would obtain a set of individual images at different tilt angles. These could then be further processed in a single-particle-like approach, which allows movie-mode drift correction and precise correction of electron microscopy artifacts (such as the CTF), so that a much higher resolution can eventually be achieved at the end of this process. In several cases, this single-particle processing of electron tomography tilt series led to side-chain-resolving resolution on proteins that were still within the cellular enviroment.